1 Introduction
Genomic medicine is an emerging medical discipline that uses the human genomic data of an individual as a part of their clinical care. Besides healthcare, genomic data is used in various domains, including research on genomic-wide association studies, ancestry determination, legal cases (e.g., paternity cases), and forensic and criminal investigations. The potential of genomics could revolutionize clinical care by providing targeted diagnostics and treatment for patients based on their genetic makeup, identify the genetic predisposition for an individual to serious disease and determine if the potential offspring may develop rare genetic disease based on genomic data of parents, etc. Thus the analysis of genomic data and development and implementation of genomic medicine based on genomic data analysis has tremendous economic potentials, which indeed explains the success of many enterprises such as 23andMe, Color Genomics and some recent services from Google Genomics, IBM Watson, Microsoft Genomics, Amazon AWS Genomics and Apple Research Kit.
Thus the recent advances in genomic research are leading to a new era in medicine. In the next few years, the use of genomic data in healthcare will rapidly increase. In the future, decisions regarding the prevention and treatment of diseases will be increasingly based on an individual’s genetic makeup. This major change in medicine requires careful preparation. Even though the promise of personalized diagnoses and treatment based on genomic data analysis and genomic medicine seems just around the corner, the study of genomics is becoming a field that is dominated by the growth of data.
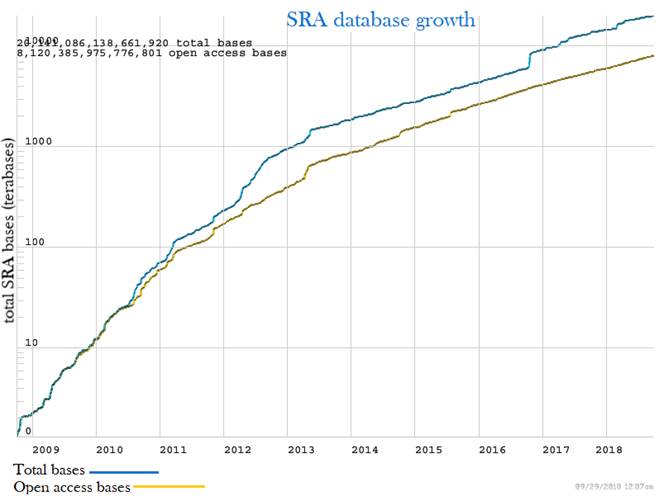
Figure 1: An overview of the publicly available data at the Sequence Read Archive (SRA) based on user-submitted metadata. The SRA’s growth rate is greatly increasing over time.
Half of the SRA submissions have been generated by genomic library strategies, mostly whole genome sequencing. The second half is composed of library strategies from transcriptome, epigenomic, and other applications. Figure adapted from (NCBI, 2018).
Below, we identify the useful collections of publicly available next-generation sequencing (NGS) datasets and how new advances in NGS technologies are greatly expanding the current volume and the range of existing data. The Sequence Read Archive (SRA) is an international public archival resource for NGS data established under the guidance of the International Nucleotide Sequence Database Collaboration (INSDC). Instances of the SRA are operated by the National Center for Biotechnology Information (NCBI), the European Bioinformatics Institute (EBI) and the DNA Data Bank of Japan (DDBJ). The mission of INSDC is to preserve public-domain sequencing data and to provide free, unrestricted and permanent access to the data. By the start of 2012, approximately 75 000 genomic, 15 000 transcriptome and 15 000 epigenomic submissions had been contributed to the SRA (figure 1). However, that volume of data represents only the tip of the iceberg as the number of genomic submissions has been steadily increasing, particularly in recent years.
Huge amounts of genomic data requires high performance computing and data storage infrastructure that is often beyond the capabilities of single institution. For this reason, Cloud computing is becoming the preferred solution for medical research centers and healthcare providers to efficiently deal with the increasing amount of genomic data in flexible and cost-effective way.
Cloud computing facilitates the storage and management of large amount of data, acts as a final destination for heavy-weight processing, long-term storage and analysis. Cloud computing eliminates the expenses of computerization and framework support and is flexible and cost-effective approach to genomic data management. Cloud computing service providers offer services that provide the infrastructure, software, and programming platforms to clients, and are accountable for the cost for development and maintenance. Challenges of using Cloud computing for genomic data include lengthy data transfers for uploading data to the cloud server, the perceived lack of information safety in Cloud computing, and the requirement for developers with advanced programming skills.
Also, since the number of devices connecting to the cloud is growing, there is undue pressure on the cloud infrastructure. Due to the loosely controlled and non-homogeneous nature of the internet there are several other issues related to Cloud computing that are still unresolved. One such issue is network delay or lag between client request and cloud response for real time applications. This can be explained by fact that data centers are often located far away from major cities and population areas. This physical distance between data centers and end users has an impact on latency which could affect the users. This delay can be a major issue for applications which rely heavily on storage, streaming data and offline processing.
Fog computing is a natural extension of Cloud computing and is foreseen as a remedy to eliminate such issues. Cloud and Fog computing share overlapping features, but Fog computing has additional attributes such as location awareness, enhanced mobility features, edge deployment, support for real-time processing (Botta et al., 2016). In contrast with centralized clouds, fog nodes are geographically distributed and deployed in large numbers near wireless access points in areas which sustain the heaviest usage, thus having a close proximity to end-users and offers a mobile, low latency, latency-sensitive analytics for mission critical requirements and real-time interaction. Rather than a substitute, Fog computing often serves as a complement to Cloud computing (Baccarelli et al., 2018). The concepts of cloud and Fog computing can be integrated into a single platform to achieve the best of both worlds: reduced latency, geographic awareness, improved data streaming, and access to commodity resource pools (Madsen et al., 2013). In table 1, we made a general overview of the existing frameworks for the Big Data (BD), Data Centers (DC) and Internet of Things (IoT) environments and highlight their advantages and disadvantages.
Table 1: Overview of BD/DC/IoT and their advantages and disadvantages in Fog Computing supports
| Frameworks | Fog Computing Support (BD + DC + IoT) | |||
|---|---|---|---|---|
| Pros | Cons | |||
| BD | ||||
| (Bonomi, 2011; Bonomi et al., 2012; Bonomi et al., 2014) (Kai et al., 2016) (Saharan & Kumar, 2015) | BD in Fog is distinguished by volume, velocity, variety and geo-distribution Applications data are passed in several layers Eliminate delays in data transfer Allows to keep data close to users instead of storing them in far DCs Reduces the time-scale in real-time | Defines policies to specific security, isolation and privacy during multi-tenancy | ||
| +DC | ||||
| (Bonomi, 2011; Bonomi et al., 2012; Bonomi et al., 2014) (Saharan & Kumar, 2015) | Degree of consistency between collection points | Policies to defined network, storage, compute with a service such as minimum delay rate etc. | ||
| +IoT | ||||
| (Bonomi, 2011; Bonomi et al., 2012; Bonomi et al., 2014) (Firdhous et al., 2014) (Byers & Wetterwald, 2015) (Saharan & Kumar, 2015) (Luan et al., 2015) (Kai et al., 2016) | Permits ordinary physical or daily life object connections Fast mobile apps Improves the QoS through local fast-rate connections Distributed intelligence Scalability Network resource preservation Close loop control Resilience Clustering | Not clear definitions of policies to specify thresholds for load balancing such as minimum number of users, connections, CPU load etc, and policies to specify QoS requirements. | ||
| 5G Technologies | Network Function Virtualization (NFV) | Software Defined Networking (SDN) | ||
| (Luan et al., 2015) | ||||
Cloud computing powerful technology to perform massive-scale/complex computing and Fog computing offers a promising solution to move computational capabilities closer to the data generated are the solution that is gaining traction in genomics research. Fog computing technology can provide a way for research to enhance their capability to store and share data, save time and reduce costs of data sharing.
This paper presents an idea of fog assisted cloud paradigm for genomic data analysis. In the next sections we introduce cloud and fog paradigms, motivation and application of fog assisted cloud paradigm to genomic data analysis and the Collaborative-Fog (Co-Fog) paradigm. We finally conclude with conclusions and future research opportunities.
2 Cloud and Fog Computing Paradigm
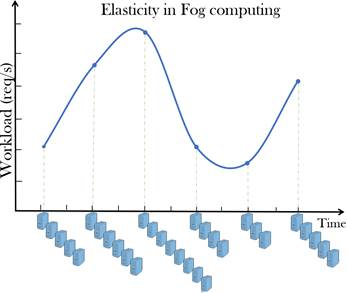
Figure 2: Elasticity of Fog computing is its ability to expand or contract its dedicated resources to meet the current demand. Elasticity is one of the feature of Fog associated with scale-out solutions (horizontal scaling), which allows for resources to be dynamically added or removed when needed. In virtualized environments Fog elasticity could include the ability to dynamically deploy new virtual machines or shutdown inactive virtual machines. The workload vs. time plot shows the elasticity of Fog computing model, where it has ability to add and remove resources “on the fly” to handle the load variation.
Cloud and Fog computing are two standing-alone technological paradigms under the real of the Future Internet. The National Institute for Standards and Technology (NIST) formally defines the Cloud computing paradigm as a model for enabling convenient, on-demand network access to a shared pool of configurable computing resources (e.g., networks, servers, storage, applications, and services) that can be rapidly provisioned and released with minimal management effort or service provider interaction (NIST, 2018a). Cloud computing provides the tools and technologies to build data/compute intensive parallel applications with much more affordable prices compared to traditional parallel computing techniques. Cloud computing offers higher-level services, unlike local server or a personal computer that can be rapidly provisioned and released with minimal management effort and relies on the self-establishment and self-management in order to guarantee scalability to large scale (Mell & Grance, 2011; Armbrust et al., 2010; Fox et al., 2009; Buyya et al., 2009). Cloud providers deliver to the users mainly three types of service models: i) Infrastructure as a Service (IaaS): IaaS providers offer to the users a pool of computing storage and network resources; ii) Platform as a Service (PaaS): PaaS providers enable users to access a platform to develop and deploy software, and; iii) Software as a Service (SaaS): SaaS users access software running on servers. Cloud computing facilitates the storage and management of large amount of data and can serve as a possible instrument of surveillance. The clouds act as the final destination for heavy-weight processing, long-term storage and analysis.
NIST in March, 2018 released a definition of the Fog computing by adopting much of Cisco’s commercial terminology as published in NIST special publication 500-325 (NIST, 2018b). Fog computing conceptual model defines Fog computing as a horizontal, physical or virtual resource paradigm that resides between end devices and Cloud computing data centers. This paradigm supports vertically-isolated, latency-sensitive applications by providing ubiquitous, scalable, layered, and federated distribution of the communication, computation, and storage resources. Precisely, fog nodes (physical (e.g. gateways, switches, routers, servers, etc.) or virtual components (e.g. virtualized switches, virtual machines, cloudlets, etc.)) are small-size virtualized inter-connected resource-equipped data centers, which are hosted by wireless access points at the edge of the network, in order to build up a two-tier FOG-CLOUD hierarchical architecture. Fog computing natively supports inter-Fog resource pooling. Furthermore, Fog computing handles data at the network level, on smart devices and on the end-user client side, instead of sending data to a remote location for processing (Bonomi et al., 2012; Horne, 2018). In Fog computing, cloud elastic resources are extended to the edge of the network, such as portable devices, smart objects, wireless sensors and other IoT devices to decrease latency and network congestion (Tang et al., 2017). In virtualized environments, Fog elasticity (figure 2) could include the ability to dynamically deploy new virtual machines or shutdown inactive virtual machines.
Next-generation sequencing is growing in number, and these data requires using large-scale computational resources. Cloud computing’s powerful technology to perform massive-scale/complex computing coupled with the Fog computing’s promising solution to move computational capabilities closer to the source of data generation thereby enhancing the capability to store and share data, save time and reduce costs of data sharing could provide an exceptional solution in the field of genomics research.
3 Motivation and Application to Genomics
Cloud and Fog computing are considered by investigators to manage and share the vast amounts of genomic data generated following NGS. It was recognized that data storage/management is a growing problem which will require Cloud and Fog technology (Charlebois et al., 2016).
For science users, Fog computing is having two main advantages: reproducibility and local/global access. Fog computing exhibits the right features for coping with the aforementioned technological issues. The Fog computing architecture consisting of three components, namely: IoT nodes, fog nodes and back-end cloud, where fog nodes are small-size virtualized inter-connected resource-equipped data centers, that process tasks without third-party interference and collaboratively provide computational flexibility, better communication, storage capacity, and much more additional new smart services in a hierarchical environment for rising number of end users in its close proximity. In addition, Fog computing natively supports three main services: 1) user virtualization; 2) User-to-Fog task offloading, and; 3) inter-Fog resource pooling. These services could be efficiently exploited, to implement the Co-Fog collaboration network as an overlay network of user clones, that entirely relies on the bandwidth/computing resources of the supporting fog nodes. The Cloud computing addressed many problems posed by data archives. But the Fog elasticity allows users to scale computing resources in proportion to the amount of data being analyzed, sidestepping constraints imposed by fog nodes. Input data can be downloaded directly to the fog nodes that will process it, where the field has produced an array of uniformly processed and summarized data-sets.
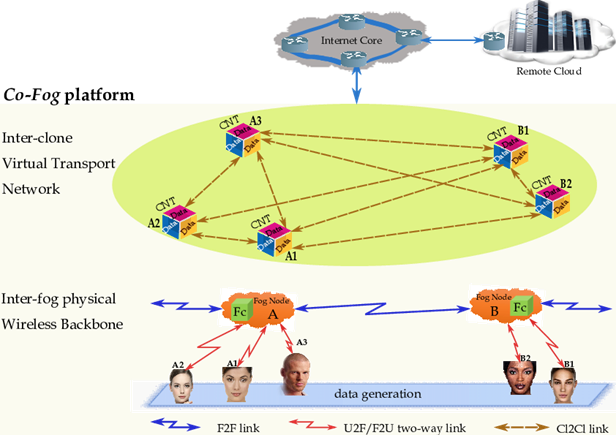
So doing, the native resources of the physical things could be employed only for the synchronization with the corresponding Fog-hosted clones, allowing to the users, perhaps on opposite ends of the globe, to create near-identical hardware and software setups (figure 3), where the reproducibility advantages are possible on non-cloud computers using Virtual Machine (VM)-based technology and the (emerging) container based technology, which are tools that package software with all the necessary components to enable reproducible deployment in different computing environments. This is indeed, the main idea behind the proposed Co-Fog paradigm. Also, Fog providers maintain data centers in such a way that achieves economies of scale and Fog users need not be concerned with outages, software patches, service contracts or damaged parts.
Passing to describe figure 3 that sketches the reference architecture of the Big Data (BD) technological platform. It is composed of five blocks, which are: 1) the IoT (data generation) layer; 2) the radio access network; 3) the proximate Fog layer; 4) the Internet backbone, and; 5) the remote Cloud layer. So, according to the reported architecture, BD streams are: i) gathered by a number of spatially heterogeneous devices (users) scattered over the environment of interest; ii) forwarded to proximate fog nodes over (single-hop) WiFi connections for local pre-processing (like, data compressing, fusion and filtering), and; iii) routed to cloud-based remote data centers over (multi-hop) internet WANs for further post-processing.
The goal is to allow users to share information in real-time by leveraging Fog-assisted social network platforms (like Globus Genomic, Dropbox, iCloud). These applications require massive sets of inter-stream cross-correlation analytics, to quickly detect the occurrence of new social trends and/or anomalies.
Fog services is comprised by three deployment models (public, private or hybrid) that vary depending on the extent to which they are freely accessible (Dove et al., 2015; Forer et al., 2012; Ruiter & Warnier, 2011; Shanker, 2012). The decisions over the type of services and deployment models influence the form Fog computing adoption and reflect how varying ethical, legal and social challenges are managed.
3.1 An Introductory Example
The definition of cloud and fog computing applied to the field of genomics research is "a scalable service where genetic sequence information is stored and processed virtually, usually via networked, large-scale data centers accessible remotely through various users and platforms over the internet" (Dove et al., 2015).
The following illustrative example (see figure 4) aims at giving some first insight into the Fog-over-Users interplay and the roles played by the Computer-to-Computer (C2C) interaction model, in order to establish collaboration, links, and ties.
In the figure 4 we show the main building blocks and involved users, where the fog fosters reproducibility by enabling investigators (users) to publish data-sets (resp., User A (Data A) and User B (Data B)) in figure 4) to the fog/cloud, including different versions thereof, without loss or modification of the previous data-sets. Moreover, the users can be situated near or far geographically (resp., User A (Data A), User B (Data B), User C (Data C), and User D (Data D) in figure 4) and can clone data-sets within the fog applied customized software to perform their own analyses and derive new results. Independent investigators can copy original/primary data-sets, softwares and published results within the fog/cloud to replicate published analyses, can be on opposite ends of the globe, to create near-identical hardware and software setups.
The Horizontal traffic Offloading (HO) capability offered by the nodes allows the implementation of the inter-clone Co-Fog network by sustaining the required C2C interactions (see the blue paths of figure 4), while the corresponding Vertical traffic Offloading (VO) capability makes feasible User-to-Clone and Clone-to-Cloud synchronization (the red paths of figure 4). The fog nodes fully support data mining, Clone-to-User communication, and inter-clone communication (the green paths of figure 4). So, it is expected that the native resources of the physical things are saved.
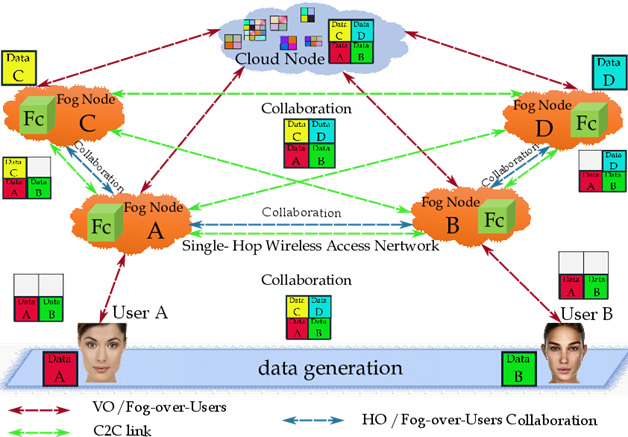
Figure 4: An illustrative example of the Fog-over-Users, Fc=Fog clone; VO= Vertical Offloading; HO= Horizontal Offloading; C2C=Computer-to-Computer.
The Fog is also accessible globally, so that, the investigator anywhere of the world can rent resources from a provider, regardless of whether the investigator is near a data center. Data can be secured and controlled by the collaborators without having to navigate by several institutions’ firewalls. The team members of fog can use the same commands to run the same analysis on the same (virtualized) hardware and software. This makes the Co-Fog an attractive venue for small/large genomics interaction/collaborations and also an important tool in the effort to promote robust sharing of genomics data (NIH, 2014; The Clinical Cancer Genome Task Team of the Global Alliance for Genomics and Health, 2017) (figures 3 and figure 4).
Moving on to give the complexity of genomics studies and the need to enroll patients in geographically dispersed study sites, collaboration on large-scale genomics sequencing projects at multiples sites is fairly common. Before the computational analyses begin, all relevant data is collected at whichever site that has the requisite computing capacity and experience required. If more than one site is to analyse the complete data-set, the data must be copied. The larger and the more decentralized the project, the more copies must be made. The collaborators at the various sites can use computers located near the data. Overall, the fog elasticity allows investigators to scale computing resources in proportion to the amount of data being analysed, avoiding restrictions imposed by local clusters. Input data can be downloaded directly to the fog nodes that will process it, without first going through a particular investigator´s cluster.
In some cases, the data may already be preloaded into a fog (e.g., the International Cancer Genome Consortium (ICGC) (ICGC, 2018) data are available from the Cancer Genome Collaboratory). If data are protected (e.g., dbGaP), it is possible that existing protocols will make it is possible to create a compliant fog-based computational setup (Nellore et al., 2016; Langmead & Nellore, 2018). The commands used to rent the cluster and run the software can be published or shared so that collaborators can do the same, avoiding inter-cluster compatibility issues. In the future, a series of studies can apply Co-Fog architecture to study large collections of publicly archived data.
4 The Proposed Co-Fog Paradigm
The introduction of the model for distributed collaboration ties in the data realm modifies the way in which users and things utilize the C2C interaction model in order to establish collaboration links ties. By design, under the C2C model, the users’ limits to set general rules, in order to define the community of collaborating smart devices. The services to be provided by the computers community and smart devices are both the producers and consumers of data and information.
4.1 Fog Interfaces with Cloud, other Fog Nodes, and Users
As aforementioned, Fog computing expands the Cloud computing functionality with more elasticity to the edge level of the core network to share same processing strategies and features (virtualization) and makes extendable nontrivial computation services.
The Fog computing architecture allows processing, networking, and storage services to dynamically transfer the data at the fog node, cloud, and users continuum. However, the interfaces for the fog to interact with the cloud, other fogs, and users, must facilitate the flexibility and dynamic relocation of the computing, storage, and control functions among these different entities. This will enable well-situated end-user assessment for Fog computing services and will also allow capable and effectual Quality of Service management (Anawar et al., 2018).
Roughly speaking, the proposed integrated Co-Fog architecture is based on the following main building blocks (figure 5): Fog-to-Cloud: Interfacing fog-to-cloud can be considered compulsory to support fog-to-cloud and vice-versa collaboration which provides back-to-back services. Fog-to-cloud interface also supports functionalities, such as: i) functions at fog to be supervised/managed with the Cloud computing ability; ii) cloud and fog which transfer data to each other for processing and comparing; iii) cloud which can decide to distribute/schedule fog nodes for allocation of the services on demand; iv) cloud and fog mutually can differentiate for better management computing services with each other, and; v) cloud which can make the availability of its services through fog-to-users. It is essential to find out which information and services should be transversely passed at fog and cloud. Regularity and granularity of such data and information should decide how fog or cloud can respond to that data.
In the figure 5, we observe Fog-to-Cloud (multi-hop) Internet connections, allowing fog clones to import/export data from/to the remote network. Fog-to-Fog: Fog nodes have pool resources functionality to support processing with each other. For example, all deployed fog nodes share their data storage, computing, and processing capability tasks with prioritized node functionality system for one or several users. Multiple fog nodes might also act together with service for backups of each other.
In the figure 5, we can see the inter-Fog backbone that provides inter-Fog connectivity and makes feasible inter-Fog resource pooling. Inter-Fog allows the hosted clones to exchange data by establishing C2C collaboration over an inter-clone overlay network relying on TCP/IP end-to-end transport connections and a set of inter-connected fog nodes (e.g., Fog Node A and Fog Node B), that act as virtualized cluster headers. These virtualized cluster headers host the clones (e.g., Fc A and Fc B) of the involved physical devices (e.g., Users (A1, A2 and A3) and Users (B1 and B2)).
The clones exploit the Fog support, in order to augment the computing-communication capabilities of the associated devices (e.g., Users (A1, A2 and A3) and Users (B1 and B2)). A commodity wired Giga-Ethernet switch provides intra-Fog connectivity. Fog-to-User: Fog-to-User interface essentially needs to allow the users to access fog services in user friendly environment, provide resources efficiently with secure ways. Figure 5 explains the interfaces of fog with cloud and users both, through hierarchically distributed Fog computing structure iteratively continuum.
In figure 5 we can see an emerging era of technology world from traditional Cloud computing towards nearly deployed Fog computing. It is also visualized which type of interface should be included in different type of era (e.g., fog-to-cloud, fog-to-fog, and fog-to-users). Also, it is pointed out that why we have a single and combined platform (Fog Computing) of these essential technologies. According to figure 5, a fog node covers a spatial area ??𝑎 (𝑚) and serves a cluster of users. The fog node can comprise of number of homogeneous quad-core Dell Power Edge-type physical servers, which are equipped with 3.06 GHz Intel Xeon CPU and 8 GB of RAM as an example. Each server may host the maximum number of Docker-type containers (Bernstein, 2014), the size of a container is usually within tens of 𝑀𝐵 (Zhang et al., 2018). Each container clones a thing (e.g., a user) and, according to figure 5, it is equipped with a virtual processor with a number of homogeneous virtual cores. Each thing (user) is associated to a software clone (e.g., a virtual avatar), that is hosted by the serving fog node.
In the figure 5, the wireless access network, that supports F2U/U2F communication through TCP/IP connections running atop IEEE802.11/15 single-hop links. A Virtualization layer: This layer allows each thing (user) to augment its limited resources by exploiting the computing capability of a corresponding virtual clone. The virtual clone runs atop a physical server of the fog node that currently serves the cloned user User-to-Clone: User-to-Clone connections allow the physical devices to synchronize the corresponding clones by exploiting the support of single-hop wireless access links.
The service models supported by the Co-Fog platform with the following two main remarks. First, since the fog nodes of figure 5 may play the two-fold role of offloading and aggregating points for the traffic generated by the underlying users, the Co-Fog paradigm is capable to support, by design, all the Up/Down Offloading, Aggregation and Peer-to-Peer (P2P) service models. Second, we stress that a main peculiar feature of the proposed Co-Fog paradigm is that the overlay network of figure 5 allows to move the implementation of the inter-user links from the device-based physical bottom layer to the clone-based virtual upper layer of figure 5.
The clone synchronizes the corresponding thing and works on behalf of it, in order to reduce thing energy consumption and provide bandwidth/computing thing-augmentation. U2F and F2U communications are guaranteed by transport (TCP/UDP-IP) connections that run atop IEEE802.11/15 up/down single-hop links (the red rays of figure 5). A (possibly, wireless) broadband backbone interconnects all the fog nodes (the blue rays of figure 5). Its role is to allow inter-clone communication among different fog nodes. Thing clones are logically inter-connected by end-to-end transport (TCP/IP) connections, to build up an overlay virtual network of inter-clones. For this purpose, intra-Fog wired Ethernet links (resp., inter-Fog backbone-supported wireless links) are used to instantiate transport-layer connections among clones hosted by a same fog node (resp., by different fog nodes). The corresponding clones establish a (bidirectional) collaboration. Afterward, the clones exchange the data provided by their associated data generation and, then, perform the required (cumbersome) mining of all available data. For this purpose, the computing capability of the hosting fog nodes is exploited. Finally, each clone returns the mined data to its owner (user) by using the communication capability of the host fog node.
In data generation layer each site has some computational resources and generate data. Analysis that require the full data sets are to be performed at multiple sites, requiring each of these sites to gather all portions of the data. As more fog nodes join to analysis, more copies must be made. Multiples fog nodes can organize themselves into a federated fog, where each analysis of the data set is automatically coordinated to minimize data transfer. The computers located where the data is generated are used to analyse that subset. Fog nodes can also consolidate their data in a cloud-based data center where the analyses are performed.
4.2 Co-Fog in Genomics
Co-Fog will be a natural fit for sharing across borders; data may be housed in the originating jurisdiction, where control is maintained, while authorized access is available outside the jurisdiction through a common interface.
Many collaborations already use the cloud to consolidate project data. ENCODE (ENCODE, 2018) uses the DNAnexus (DNAnexus, 2018) platform for cloud-based analysis and data sharing, and DNAnexus in turn uses the infrastructure of AWS (Genomics, 2018) modENCODE (modENCODE, 2018) and ICGC (ICGC, 2018) where both host their data sets in the cloud through AWS.
With Co-Fog the collaboration can also be implemented to the above given example which solely using cloud. By using Co-Fog platform which encourages another dimension of strength borrowing, it will be easier to leverage public data. The investigators can use public data to boost the power available to analyse a locally generated data set, a paradigm that can prevails in microarray data analysis. We expect Co-Fog can be used for running large-scale analysis across multiple fogs, even to the point that new sequencing data analyses can be performed in the Fog and with the benefit of being able to see across many studies with important variables in common.
As an example, RNASeq-er enables investigators who have submitted unpublished sequencing data to the ArrayExpress archive to automatically analyse that data, free of charge, using computational resources at the EMBL-EBI in the context of other public data in the archive. When the study is published and the data become public, the summarized results can be joined with those of the other published, archived studies. Thus, the investigator benefits before publication and the community benefit after (Langmead & Nellore, 2018).
4.3 The Container-based Virtualization Technology
Virtualization is employed in Fog-based data centers, in order to (Baccarelli et al., 2017):
• dynamically multiplex the available physical computing, storage and networking resources over the spectrum of the served devices;
• provide homogeneous user interface atop (possibly) heterogeneous served devices; and,
• isolate the applications running atop the same physical servers, in order to provide trustworthiness.
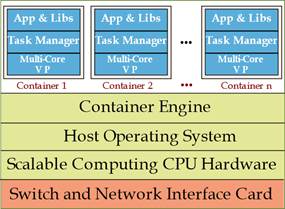
Figure 6: Container-based virtualization of a physical server equipping a Fog node (Virtualized server architecture); VP=Virtual Processor.
Roughly speaking, in virtualized data centers, each served physical device is mapped into a virtual clone that acts as a virtual processor and executes the programs on behalf of the cloned device (Portnoy, 2012). In principle, two main virtualization technologies could be used to attain device virtualization, namely, the (more traditional) Virtual Machine (VM)-based technology and the Container based technology (Bernstein, 2014; Soltesz et al., 2007).
According to figure 6, a virtualized physical server running at a fog node is composed of: i) containers where each container plays the role of virtual clone for the associated physical thing. Hence, the container acts as a virtual processor and executes the tasks offloaded by the thing on behalf of it. For this purpose, it is equipped with a Virtual Processor (VP), that runs at a (scalable) processing frequency 𝑓(𝑏𝑖𝑡/𝑠) and it is controlled by a Task Manager. The VP executes the programs stored by the corresponding Application Library (figure 6), all the application libraries stored by the instantiated containers must be compliant with the Host Operating System (HOS) equipping the host physical server; ii) the pool of computing (e.g., CPU cycles) and networking (e.g., I/O bandwidth) and physical resources made available by the CPU and Network Interface Card (NIC) that equip the host server.
Container Engine dynamically multiplexes the resources of the fog server over the set of hosted containers, and; iii) a HOS which is shared by all hosted containers (figure 5)(Baccarelli et al., 2017).
Due to the expected large number of devices (users) to be virtualized, resorting to the container-based virtualization would allow to increase the number of virtual clones per physical server (e.g., the so-called virtualization density) (Xu et al., 2014).
5 Conclusion and Future Work
Cloud computing is changing how large computational resources are organized and acquired. It is also changing how scientists and researchers in genomics collaborate and deal with vast archived data sets. However, Fog computing provides certain advantages over Cloud computing like faster data processing with reduced latency, location-based customization, etc. However, Fog computing is not a replacement for cloud computing as cloud computing will still be desirable for high end data storage, analysis and processing jobs in scientific fields like genomics.
We adopt Fog computing as a necessary tool for advance genomic research to collaborate and deal with vast sets of archived data, so that the genomic investigators would be able to do immediate collaborations.
As more archived data will be housed in the fog, where the Fog computers which are physically proximate to the generated data can access it rapidly and will allow new modes of analysis, interaction and collaboration.
Co-Fog is the ways to address the crucial issues that arise as the scientists increasingly use large-scale genomics data to improve our understanding of biology and disease. Also, considering the genomic data to be user sensitive, there will be a need for genomics investigators to understand the fog and also remain responsible for their data stored in the fog.
The proposed (Co-Fog) model has advantages of higher speed, greater accessibility and collaboration over Cloud computing which makes it a preferred model for genomic data analysis, however, Co-Fog can be applied in several other research field involving Big data storage and analysis. Since, Co-Fog relies on the distributed networked computing architecture, its innovative solutions are expected to successfully tackle the issues in distributed security, in order to allow the migration of the Co-Fog paradigm from the theory to the practice.