Introduction
In recent years, there have been significant advancements in the treatments of neuropsychiatric diseases. The first psychotropic drugs' appearance has been conceived to treat most neuropsychiatric conditions such as bipolar disorder (BP), schizophrenia (SP), mania, hypomania, to name a few. Even though these drugs' benefits are well known, the variation on treatment efficacies shown among individuals has become a topic of great controversy regarding their use (1). According to the literature, most neuropsychiatric patients continue to experience symptoms related to their conditions even after treatment, with a minority going into remission (1)(2). Researchers on this matter have focused on understanding the mechanism behind such low efficacy of the drugs by unveiling genetic factors. An intensive research area has focused on understanding how genetic polymorphisms in biological receptors used for these drugs might influence their pharmacokinetic profile (3)(4). The importance of such studies has led to the emergence of two new fields, the first one known as pharmacogenetics, which correlates the differences in drug response among individuals due to single genetic mutations, and the second one known as pharmacogenomics, which studies the drug response based on different genetic activity across the genome of an individual (5).
These two fields extensively explore the treatment response of neuropsychiatric conditions. Studies have been driven by low remission rate of such conditions and the lack of understanding of their etiology (6). These problems are related to the treatment with antidepressants, antipsychotics, and mood stabilizers. For example, in depressed patients, it was reported that 30-40% of the individuals did not show any treatment response with antidepressants (7)(8)(9). This condition was observed even at six weeks after the treatment was commenced. A similar observation was outlined to treat bipolar disorders with lithium, where a partial or total response was only reported for 70 to 80% of the patients (10)(11). This situation is not different from the treatment response of antipsychotics used to treat conditions such as schizophrenia, where up to 30% of patients were not successfully treated (12)(13). This evidence has made possible a continuous increment in pharmacogenomics and pharmacogenetics studies in patients suffering from mental health conditions.
Indeed, most research studies have focused on correlating the drug response to specific genes linked to an enzyme and protein functions or biochemical pathways that could influence drug metabolism. In this regard, the cytochrome P450 enzymatic system is the most studied in treating neuropsychiatric diseases (14)(15)(16)(17). Some studies suggest linked polymorphisms in the CYP450 family and the treatment of adverse effects (18). Other researchers, however, have been looking for genes that correspond to specific receptors in the brain. Some interesting published works on this matter are genome-wide association studies in response to antidepressants (19)(20) or lithium (21). It was found that genes including UBE3C, RORA, BMP7, and some single nucleotides polymorphisms (SNP) might influence how a patient responds to treatment with antidepressants, whereas genes such as ODZ4, SDC2, and GRIA2 might be responsible for lithium response.
Studies on an individual's drug response based on his/her genetic profile have had great relevance towards the advancement of personalized neuropsychiatry. However, the lack of homogeneity among findings and the significant number of variables that should be considered for these studies have diminished clinical relevance. One strategy that could be used to increase the homogeneity and, hence the relevance of the studies, is to classify the mixed results in terms of ethnicity. Due to the available human genome sequencing methods, the research on diseases that affect specific racial groups has improved dramatically (22).
In this study, a systematic review was done employing the PRISMA methodology, which aimed to categorize the vast amount of information found for neuropsychiatric disorders: bipolar disorder (BP), schizophrenia (SP), mania, hypomania, and major depressive disorders (MD) and how specific treatment responses are related to ethnicity. PRISMA is an evidence-based minimum set of items for reporting in a systematic review. For this, we considered pharmacogenetics and pharmacogenomics studies on drug response for antidepressants, lithium, anticonvulsant, and various antipsychotics for treating each condition. Ultimately, this review might serve as a concise work for future research on pharmacogenetics and pharmacogenomics treatment of neuropsychiatric conditions in specific ethnic groups.
Methods
Eligibility criteria:
a. All studies in pharmacogenetics and pharmacogenomics
b. Citing ethnicity and ethnic groups
c. In patients with bipolar disorder (BP), schizophrenia (SP), mania, hypomania, and major depressive disorders (MDD)
d. In treatment with all classes of antidepressants and antipsychotics.
Information sources: An electronic search was performed on PubMed, Medline, Web of Science, Scopus, and Google Scholar, considering any studies published in that database at any time until November 30th, 2019.
Search: We used the following terms: pharmacogenetics, pharmacogenomics, ethnicity, and ethnic groups combined with search terms from the treatment (antidepressants, lithium, anticonvulsant, and various types of antipsychotics) and the condition (bipolar disorder (BP), schizophrenia (SP), mania, hypomania, and major depressive disorders (MDD). We searched in each database; an input contained terms as pharmacogenetics OR pharmacogenomics AND ethnicity OR "ethnic groups" AND "bipolar disorders" AND "lithium."
Study selection: We transferred all the studies to a table where we analyzed using both the title and the abstract. We excluded a study if it did not meet the inclusion criteria. Some studies encountered in the search mainly focused on the treatment's adverse effects. Other studies discussed some individuals' susceptibility to suffer psychiatric disorders due to a specific polymorphism, therefore rendering them ineligible. We eliminated all duplicated studies within the sub-search. If similar studies were found in two sub-searches, they were kept, and priority was given based on the search's relevance.
Data collection process: a single author extracted all data. We validated the search methodology by a peer-review process. The paper selection, however, was performed individually. We considered primary research articles, as well as review studies. In the reviews, if it was necessary, the primary article was consulted (23). For them, key aspects were looked at to narrow further the results, such as the study's significance, the strong correlation of the polymorphism, and differential drug response. A flowchart of a model search in a database can be found in Figures 1, 2, and 3. (Figure 1)(Figure 2)(Figure 3)
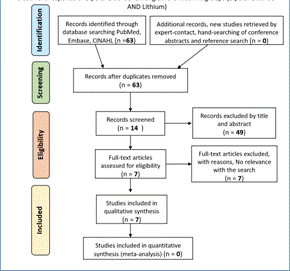
Figure 1 Search strategy for the study of pharmacogenetics and pharmacogenomics in the treatment response of Bipolar Disorder among different ethnic groups (Bipolar Disorder AND Lithium)
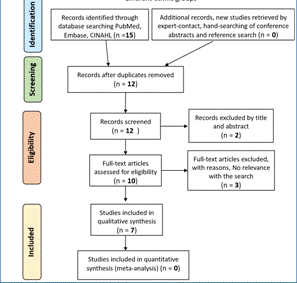
Figure 2 Search methodology for the study of pharmacogenetics and pharmacogenomics in the treatment response of Schizophrenia among different ethnic groups.
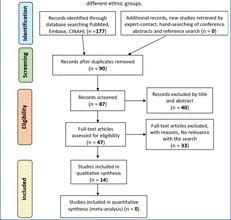
Figure 3 Search methodology for the study of pharmacogenetics and pharmacogenomics in the treatment response of Major Depressive Disorder, Mania o Hypomania among different ethnic groups.
Data items: Data extracted from each study included: authors, year of publication, study purpose, drug group, drug name, gene polymorphism, reported association, effect size, Odds Ratio, number of individuals analyzed, ethnicity.
Risk of bias in individual studies: We discussed the risk of bias at the outcome level.
Synthesis of results: Based on our evaluation of publications to date, it is clear that the level of heterogeneity across the studies is relevant in terms of study purpose, drug group, drug name, Gene Polymorphism, reported association, effect size, Odds Ratio, number of individuals analyzed, and ethnicity. We synthesized data with information provided in the text and tabular form to summarize the review's findings.
Results
Study selection: the electronic search for schizophrenia and antipsychotics yielded 154 studies, including duplicates. We excluded most of these articles (69.48%) because they did not meet the criteria based on title and abstract reviews. We excluded 33 articles from the full-text review because their focus was a genetic predisposition to develop schizophrenia. Thus, only 14 articles were included (24)(25)(26)(27)(28)(29)(30)(31)(32)(33)(34)(35)(36)(37). For major depressive disorder, following a similar screening method 22 initial studies were narrowed down to 7, which were included (38)(39)(40)(41)(42)(43)(44). Interestingly, the search for mania or hypomania in relation to drugs such as lithium, anticonvulsant, and antipsychotic resulted in four articles that were all excluded.
Study characteristics: As seen in Table 1, genes studied were BDNF, PTPRD, CYP2D6, HTR2A, TPH1, SCL18A1, COMT, DRD1, DRD2, DRD3, 5HTT, D2, DAT, H5T6, ITIH3, CCL2, HTR1A, HTR3B, CYP1A2, ABCB1. Genetic studies on CYP2D6, HTR2A, COMT, DRD1, DRD3, and D2 were repeated across some of the articles found. However, the polymorphisms studied for drug response were different. Only one polymorphism was repeated (rs4532) across several studies on the DRD1 gene. Overall, from the 14 articles, 26 ethnic groups were included: 10 featured participants from European/Caucasian origins, 4 on Chinese, 1 on Korean, 4 African, 2 Russian, 2 Tatar, and 1 Indian. From the analysis of these studies, no significant association was found on 3774 Europeans between Val66Met polymorphism on BDNF gene and treatment with olanzapine, risperidone, or quetiapine (24). Similarly, no significant correlation was found between 1846G>A on CYP2D6 gene and clinical response to either typical (haloperidol) or atypical (Risperidone, Paliperidone, Quetiapine) drugs (30). No association was found in the Asian population in 185 Han Chinese people with rs5326 polymorphism in the DRD1 gene and treatment with risperidone (26). In a comparative study, rs265976 DRD1 gene was associated with positive symptoms after Clozapine treatment on 49 African Americans, which was not observed on Caucasians (32). One study identified that side effects of some drugs varied with different polymorphisms on the PTPRD gene. These responses might include weight gain after clozapine or olanzapine treatment on Europeans (rs7339824) and African Americans (rs7040189). The COMT gene with three polymorphisms (rs6269, rs5993883, rs4818) showed a significant association with treatment response (37). Other polymorphisms, such as the Val108/158Met, were associated with cognitive improvement (34) and rs165599, was associated with sensitivity development (31). (Table 1)
Table 1 Current studies on pharmacogenetics of schizophrenia and ethnicity.
| Study reference | Drug group | Drug name | Gene | Polymorphism | Reported association | Effect size OR | n= | Ethnicity |
|---|---|---|---|---|---|---|---|---|
| (24) | Atypical | Olanzapine Risperidone Quetiapine | BDNF | Val66Met | No significant association | 1 | 3774 | European |
| (25) | Atypical | Clozapine or Olanzapine | PTPRD | rs10977144 | No effect on weight gain | NA | 144 | European |
| rs73398242 | Associated with weight gain | NA | ||||||
| rs10977154 | No effect on weight gain | NA | 57 | African American | ||||
| rs7040189 | Associated with weight gain | NA | ||||||
| (30) | Typical | haloperidol | CYP2D6 | 1846G>A | No significant association | NA | 37 | Russian |
| Atypical | Risperidone paliperidone Quetiapine | NA | 42 | Tatar (Turkic ethnic group) | ||||
| (31) | Atypical | Clozapine | HTR2A | rs799012 | Development of sensitivity | NA | 128 | Russians and Tatars |
| rs6311 | ||||||||
| rs614 | ||||||||
| Typical | Haloperidol | TPH1 | rs1800532 | |||||
| SCL18A1 | rs2270641 | |||||||
| COMT | rs165599 | |||||||
| (32) | Atypical | Clozapine | DRD1 | rs265976 | Positive symptoms | NA | 49 | African American |
| rs4532 | No association | 183 | European | |||||
| (33) | Atypical | Clozapine or Olanzapine | DRD2 | rs6277 | Associated with weight gain | 3.37 | 85 | European American |
| rs180049 | 2.18 | |||||||
| (34) | Atypical | Clozapine | COMT | Val108/158Met | Cognitive improvement | NA | 59 | European |
| 25 | African | |||||||
| DRD3 | Ser9Gly | Higher frequency of gly allele in responders | 1.4 | 32 | Pakistani | |||
| 5HT2A | 102-T/C | C/C patients had poor response | 3.4 | 149 | European | |||
| -1438-G/A | G/G individuals had poorer response | 2.4 | 144 | European | ||||
| 40 | African American | |||||||
| Olanzapine | 5HTT | LRP | Better treatment response | NA | 94 | Caucasian | ||
| Risperidone | D2 | Ser311Cys | Homozygotes showed a poorer response | NA | 123 | Asian | ||
| (35) | Atypical | Nemonapride, Bromperidol, Chlorpromazine | D2 | -141C Ins/Del | Improvement with anxiety | NA | 49 | Asian |
| Olanzapine | DAT | VNTR | No association | 75 | European | |||
| Clozapine | H5T6 | 267-T/C | Better response | 99 | Asian | |||
| (36) | Atypical | Clozapine | ITIH3 | rs2535629 | Improvement of negative symptoms | NA | 256 | European |
| (37) | Atypical | Quetiapine | COMT | rs6269 | Treatment response | 1.938 | 995 | Asian |
| rs5993883 | 1.877 | |||||||
| rs4818 | 0.504 | |||||||
| (26) | Atypical | Risperidone | DRD1 | rs5326, | No association | NA | 185 | Asian |
| rs4867798 | ||||||||
| rs4532 | ||||||||
| rs686 | ||||||||
| (27) | Atypical | Risperidone | CCL2 | rs4795893, | Treatment associated | NA | 208 | Asian |
| rs1024611 | ||||||||
| rs4586 | ||||||||
| rs2857657 | ||||||||
| (28) | Atypical | Risperidone, Olanzapine, Clozapine, Ziprasidone, Quetiapine, Aripiprazole, and Amisulpride | DRD1 | rs265967 | Nonresponder to atypical treatment | NA | 371 | Asian |
| DRD3 | rs10934254 | |||||||
| HTR1A | rs878567 | |||||||
| HTR3B | rs1176744 | |||||||
| (29) | Atypical | Norclozapine and clozapine | CYP1A2 | rs2069521 | No association | NA | 96 | Asian |
| rs2069522 | ||||||||
| CYP2D6 | rs1135840 | |||||||
| Clozapine | ABCB1 | rs7787082 | Associated with non-responders | 4.87 | ||||
| rs10248420 | 4.1 |
*Elaboration: authors
With the search for pharmacogenetics and major depressive disorder 10 genes were found, 5-HT2A, HTR2A, GRIK4, SLC6A4, ABCB1, NTRK2, SLC6A2, BDNF, PDE11A, PDE9A (Table 2). None of the genes were studied in more than one article. Some genes such as BDNF, HTR2A, and ABCB1 were also related to treatment response for schizophrenia. rs24531 polymorphism on SLC6A4 was the only one reported as having no association between treatment with fluoxetine and major depressive disorder.
Table 2 Current studies on pharmacogenetics of mania as well as major depressive disorder and ethnicity.
| Study reference | Drug group | Drug name | Gene | Polymorphism | Reported association | Effect size OR | n= | Ethnicity |
|---|---|---|---|---|---|---|---|---|
| (38) | SSRI | Paroxetine or fluvoxamine | 5-HT2A | A-1438G or T102C | Slower score in somatic anxiety | NA | 203 | European |
| (39) | SSRI | Citalopram | HTR2A | rs7997012 | Association with treatment response and remission | NA | 1850 | European |
| GRIK4 | rs1954787 | |||||||
| (40) | SSRI | Fluoxetine | SLC6A4 | rs24531 | No association | NA | 150 | Hispanic |
| (44) | TCA and SSRI | Fluoxetine and Desipramine | Do not harbor any gene | exm-rs1321744 | Significance for treatment remission | NA | 232 | Mexican American |
| (42) | TCA and SSRI | Fluoxetine and Desipramine | ABCB1 | rs2214103 | Reduction on Hamilton Depression Rating | NA | 536 | Mexican |
| NTRK2 | rs9969765 | |||||||
| rs2289657 | ||||||||
| SLC6A2 | NT_010498.15 _9300464 | |||||||
| (43) | TCA and SSRI | Fluoxetine and Desipramine | BDNF | rs61888800 | Better response to treatment | NA | 272 | Mexican American |
| SNP56133711 | ||||||||
| (41) | TCA and SSRI | Fluoxetine and Desipramine | PDE11A | rs3770018 | Treatment response | NA | 284 | Mexican American |
| PDE9A | rs729861 |
*SSRI= Selective serotonin reuptake inhibitors, TCA: Tricyclic antidepressant, NA: non-available. Elaboration: authors
Results for the pharmacogenetics of BP and ethnicity can be found below in table 3. Many of the studies were not considered for reasons similar to those explained in the other searches. Mainly because those studies focused on developing the psychiatric condition, other studies were screened out as they did not focus on treatment response but common side effects. In addition to this, most of them were neither relevant to this search, as they mentioned several other conditions. (Table 3)
Table 3 Current studies on pharmacogenetics of bipolar disorders and ethnicity.
| Study | Drug group | Drug name | Gene | Polymorphisms | Reported association | Effect size (OR) | n= | Ethnicity |
|---|---|---|---|---|---|---|---|---|
| (45) | Antimanic | Lithium | TPH2 | rs4570625 | No association | N/A | 132 | Asian & European |
| rs10748185 | No association | N/A | ||||||
| rs11179027 | No association | N/A | ||||||
| rs4469933 | No association | N/A | ||||||
| rs17110747 | No association | N/A | ||||||
| (52) | Antimanic | Lithium | TMCC1 | s2811332 | No association | 7.29 | 52 | Sardinian (European) |
| GNPDA2 | rs1390913 | No association | 12.83 | |||||
| RASSF4 | rs869156 | No association | 7.24 | |||||
| ACCN1 | rs11869731 | Better treatment response | 12.83 | |||||
| (51) | Antimanic | Lithium | GADL1 | rs17026688 | Better treatment response | 111.87 | 294 | Asian |
| rs17026651 | Better treatment response | 3.88 | ||||||
| (23) | SSRI | Fluvoxamine | 5-HTTLPR | l/l (variant) | Better treatment outcome | N/A | 53 | European |
| SSRI | Paroxetine | 5-HTTLPR | l/l (variant) | Faster treatment outcome | N/A | 58 | European | |
| SSRI + Antimanic | Fluvoxamine + Lithium | 5-HTTLPR | 16D/16A (variants) | Better treatment response | N/A | 228 | European | |
| SSRI + Antimanic | Fluvoxamine or Paroxetine + Lithium | HTR2A | 102T/C | No association | N/A | 408 | European | |
| 1420C/T | Better treatment response | |||||||
| (46) | Atypical psychotic | Clozapine | HTR-5 | A12T | No association | N/A | 105 | European |
| 19G/C | ||||||||
| (54) | SSRI | Mirtazapine | AUTS2 | rs12698828 | Better treatment response | N/A | 711 | Asian |
| rs7785360 | N/A | |||||||
| (55) | SSRI | Milnacipran or Fluvoxamine | BNDF | Val66Met | Favorable treatment response | N/A | 134 | Asian |
| (56) | SSRI | Fluvoxa mine | BNDF | RS7103411 | Worsen Treatment response | N/A | 268 | European |
*Elaboration: authors
In each independent search, the selection of the studies was performed slightly differently. For example, drug terms were searched separately, such as lithium, antidepressants, antipsychotic, and anticonvulsant, with their pharmacogenetics for treating bipolar disorders. From the results, it was observed that most of the studies focused on Asians and Europeans. Among them, the most studied gene was 5-HTTLPR (23), followed by BNDF. The gene with the most significant number of polymorphisms studied was TPH2 (45). Unfortunately, studies done on a common gene or polymorphism for all treatments were not found.
No association was found between polymorphisms in the genes such as TPH2 (45), HTR2A (23), and HTR-5(46) and their respective choice of treatment. The study with the highest number of subjects was done on an Asian cohort, and it reported a better treatment response of two polymorphisms in the gene AUTS2 to mirtazapine. Most studies considering mental health problems and their pharmacogenetics were done on many ethnicities other than Latin American or Hispanic, thus, suggesting a lack of pharmacogenetic studies in the region.
Discussion
Our study investigated the association between polymorphisms on different genes and pharmacogenetics' relationship to identify genetic determinants for drug activity. Geographical areas, ethnic groups and ethnicity influence the outcomes after drug treatment. However, these factors have often not been included in pharmacogenetic studies (47). Schizophrenia is a severe chronic mental disorder that has negative, positive, cognitive, and mood symptoms. Treatment includes medication to reduce the severity and frequency of psychotic episodes by using a known drug class called antipsychotics. These are classified into two groups, first-generation or typical agents and second-generation or atypical agents. Specific drugs reduce positive symptoms such as hallucinations or delusions and have minimal effect on negative symptoms (48).
Patients with schizophrenia might have a higher risk of developing obesity and metabolic illness such as diabetes, hypertension, and dyslipidemia, associated with their lifestyle, as well as the treatment and care received (49). This associated risk adds to the burden of this type of patient. Three polymorphisms were found to be related to weight gain after atypical antipsychotic drugs were received, two on PTPRD gene rs73398242 (Europeans), rs7040189 (African Americans) (25), and one on DRD2 gene rs6277 (Europeans) (33). Thus, to control obesity and weight gain, these different polymorphisms should be taken into consideration.
On the other hand, it was found that the 1846G>A variation on the CYP2D6 gene was standard on Europeans; typical and atypical drugs were used, but no association with treatment outcome was reported (30). These drugs' pharmacological activity differs according to their metabolites; for example, risperidone is converted to an active 9-OH-risperidone metabolite by CYP P450 2D6 hydroxylates. If inhibitors are used against this enzyme, they reduce risperidone concentration but not metabolites. Nucleotide changes on serotonergic and dopaminergic genes such as HTR2A, TPH1, SCL18A1, and COMT were reported to affect Russians' treatment sensitivity (31) significantly. A novel marker rs265976 on DRD1 was found on African Americans that had an association on positive symptoms and treatment with clozapine, in contrast to the marker found on Caucasian rs4532, which has no association with treatment (32).
Furthermore, five out of the twenty genes reported in this analysis were involved in better treatment response with atypical drugs, including LRP polymorphism in 5HTT gene; 267-T/C in H5T6; rs6269, rs5993883, rs4818 in COMT; rs4795893, rs1024611, rs4586, rs2857657 in CCL2. Even though patients with schizophrenia have an altered cytokine production, some researchers studied for the first time the genes involved in this cytokine production and the relationship with treatment response. However, most of them concluded that further research needs to be done.
While the main drug types to treat schizophrenia are antipsychotics, they are efficient only up to 30% (31). One possible explanation can be the limited existent information that relates ethnicity, race, and ancestry with drug treatment for this mental disease. All polymorphisms on the reported gene differ in each population, and none of the studies were based on Latin patients. This only confirms the need to maximize research focused on the patient's ancestry to improve their clinical outcome.
Another mental illness reviewed in this article is major depressive disorder; four out of seven articles were based on Hispanic patients and resulted in different genes with various polymorphisms. No association was found between SLC6A4 (rs24531) and fluoxetine treatment response whereas ABCB1 (rs2214103), NTRK2 (rs9969765, rs2289657), SLC6A2 (NT_010498.1_9300464), BDNF (rs61888800, SNP56133711) and PDE11A (rs3770018) resulted in better treatment response.
In terms of bipolar disorder, lithium has become the treatment of choice. Lithium has proved to work effectively in most patients to treat maniac disorders; depressive lapses are commonly observed in BP. However, some requirements need to be fulfilled to administer lithium to a patient (50). Thus, some studies focus on lithium’s action mechanism and relating it to some individuals’ genetic makeup. In this search, TPH2, TMCC1, GNPDA2, RASSF4, ACCN1, and GADL1 were genetic targets to find the association with better treatment response. From which, only the last two showed some significant association with that outcome. One of these studies examined the effect of glutamate decarboxylase-like protein 1 (GADL1) altered by specific polymorphism on lithium response in an Asian cohort.; it was found that the treatment's outcome can be predicted in 93% of the cases by studying two polymorphism, rs17026688 and rs17026651 (51). In the case of the ACCN1 gene, the authors made a whole-genome scan and found that the SNPs (rs11869731) of the ACCN1 gene can be used as a pharmacogenetic marker to? the treatment's response to lithium (52).
At this point, despite its proven efficacy, lithium has not been used as expected. More studies on understanding how lithium can improve the outcome of some mental conditions are needed. These can reduce some of the skepticism around its usage. In some countries, lithium has been suggested as their first choice of treatment due to the fear induced by some physicians. In others like Ecuador, conflicts of interest have driven such lack of usage (53). Generating knowledge can be helpful to confirm the efficacy of lithium.
Apart from lithium, drugs classified as selective serotonin reuptake inhibitors have also been used to treat BP conditions. 5-HTTLPR, HTR2A, AUTS2, and BNDF were found as target genes to explore this type of drugs' effect. Patients with some variants of the 5-HTTLPR, which encodes for serotonin transporters, have shown to recover better and faster by using this type of medication. More specifically, fluvoxamine and paroxetine (23). These studies were performed in European cohorts. Similar results were found when studying other genes such as HTR-5, AUTS2, and BNDF, where drugs like clozapine, mirtazapine, and milnacipran showed favorable treatment outcomes for these genes. An adverse outcome was found when studying the BNDF-SNPs (rs7103411)(54)(55). This result suggests that more clinical research is needed before specific treatments can be deemed highly effective (56). This is because, from the data shown, it can be seen that two SNPs polymorphism in one specific gene can trigger a different clinical response and outcome, despite having the same mechanism of action.
Conclusion
We found only five studies on pharmacogenetics of mania and major depressive disorder linked to ethnicity that involve the genes SLC6A4, ABCB1, NTRK2, SLC6A2, BDNF, PDE11A, and PDE9A, and the use of fluoxetine and desipramine. Some studies suggested that the precision of using SNPs as pharmacogenomics markers of treatment outcome, if appropriately used, could represent a significant advance in personalized medicine as two SNPs found in the same gene can yield different treatment outcomes. One expected result was that the vast amount of information found mainly reflected studies performed in European and Asian cohorts. Studies in Latino or Hispanic ethnic groups are very few, which constitutes a strong bias when choosing the appropriate drug in the treatment of neuropsychiatric diseases. Knowledge generated on the neuropharmacogenetic response to drugs can be helpful to understand any ethnic influence on drug responses.